01 Nov Defective Viral Genomes May Indicate Greater Flu Virus Severity
MedicalResearch.com Interview with:
Ana Falcón
Department of Molecular and Cellular Biology
National Center for Biotechnology
Spanish National Research Council (CNB-CSIC)
Madrid, Spain
MedicalResearch.com: What is the background for this study?
Response: Influenza A virus (IAV) infection can be severe or even lethal in toddlers, the elderly and patients with certain medical conditions. Infection of apparently healthy individuals nonetheless accounts for many severe disease cases and deaths, suggesting that viruses with increased pathogenicity co-circulate with pandemic or epidemic viruses.
IAV virulence and pathogenesis are dependent on complex, multigenic mechanisms involving the viral genetic characteristics, the host conditions, the virus-host interactions, and the host response to the infection. Influenza virus pathogenicity has been studied in depth for many years, and several amino acid changes have been identified as virulence determinants, however, a general pathogenicity determinant has not been characterized.
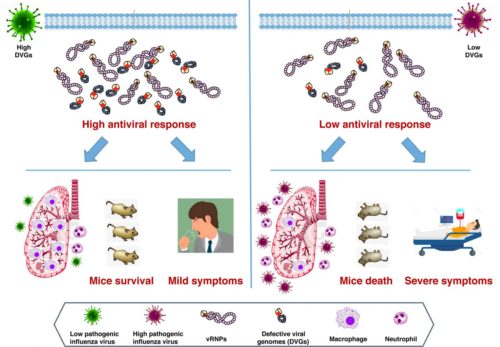
A proportion of influenza virus particles have defective genome RNAs (Defective Viral Genomes-DVGs) due to internal deletions of viral segments. The DVGs have the 3’ and 5’ ends of the parental RNA segments, and most have a single, large central deletion that generates viral RNAs of 180–1000 nucleotides. The presence of DVGs potentiates the host response in cultured cells and in animal models and leads to attenuated infection, possibly through recognition of double-stranded RNA by receptors that activate antiviral signaling cascades.
MedicalResearch.com: What are the main findings?
Response: Here we used next-generation sequencing (NGS) to evaluate the role of defective viral genomes (DVGs) in the pathogenicity of influenza virus circulating in the human population. We have identified a polymerase PA D529N mutation detected in a fatal IAV case, whose introduction into two different recombinant virus backbones, led to reduced defective viral genomes (DVGs) production. This mutation conferred low induction of antiviral response in infected cells and increased pathogenesis in mice. We found that the low amount of DVGs accumulated in tissue culture cells correlates with increased pathogenicity in mice both, in natural isolates or recombinant viruses. To analyze the association between low DVGs production and pathogenesis in humans, we performed a genomic analysis of viruses isolated from a cohort of previously healthy individuals who suffered highly severe IAV infection requiring admission to Intensive Care Unit and patients with fatal outcome who additionally showed underlying medical conditions. These viruses were compared with those isolated from a cohort of mild IAV patients. Viruses with fewer DVGs accumulation were observed in patients with highly severe/fatal outcome than in those with mild disease. We suggest that low DVGs abundance has a major role in the severe outcome of IAV-infected patients.
In summary, we establish a significant association between low DVGs accumulation and an increase in severe or fatal outcome in human influenza virus infection; we provide genetic support for this association in infected cultured cells and in mice. In addition to the previous reports about the role of DVGs in natural animal infections here we present data indicating that a reduced accumulation of DVGs may be considered a new virulence marker for viral pathogenicity in humans.
MedicalResearch.com: What should clinicians and patients take away from your report?
Response: Influenza A viruses are the causative agents of annual epidemics, sporadic zoonotic outbreaks and occasionally pandemics. Worldwide, acute respiratory infections caused by influenza A viruses continue to be one of the main causes of acute illness and death. The appearance in 2009 of a new H1N1 pandemic influenza strain reinforced the search to identify viral pathogenicity determinants for evaluation of the consequences of virus epidemics and potential pandemics for human health. Here we identify a new general virulence determinant found in a cohort of severe/fatal influenza virus-infected patients, a reduced accumulation of viral defective genomes. These molecules are incomplete viral genome segments that activate the innate immune response. Evaluation of DVGs phenotype of circulating viruses might predict its potential to induce severe disease. This data will contribute to the prediction of influenza disease severity, to improved guidance of patient treatment and will enable the development of risk-based prevention strategies and policies.
MedicalResearch.com: What recommendations do you have for future research as a result of this study?
Response: Additional work is needed to define specific DVGs function and the mechanism by which they are produced in humans. Analysis of DVGs in different influenza subtypes and viruses circulating in former years will determine the scope of this finding.
MedicalResearch.com: Is there anything else you would like to add?
Response: This finding implies a new concept on virulence determination. Regardless the mutations responsible, reduced accumulation of DVGs, constitutes a virulent factor itself. Thus, our results indicate that quantitative determination of these RNA molecules (DVGs), and not identification of specific aminoacids, might be considered as a prognosis marker for severe influenza disease.
MedicalResearch.com: Thank you for your contribution to the MedicalResearch.com community.
Citation:
Jasmina Vasilijevic, Noelia Zamarreño, Juan Carlos Oliveros, Ariel Rodriguez-Frandsen, Guillermo Gómez, Guadalupe Rodriguez, Mercedes Pérez-Ruiz, Sonia Rey, Isabel Barba, Francisco Pozo, Inmaculada Casas, Amelia Nieto , Ana Falcón
Published: October 12, 2017 PLOS Pathogens
Note: Content is Not intended as medical advice. Please consult your health care provider regarding your specific medical condition and questions.
[wysija_form id=”1″]
Last Updated on November 1, 2017 by Marie Benz MD FAAD