
09 Sep Digital Droplet Technology Can Reduce False Negative COVID-19 Tests
MedicalResearch.com Interview with:
Claudia Alteri, PhD
Assistant Professor (RTD-B) in Microbiology And Clinical Microbiology
Department of Oncology and Hemato-Oncology
University of Milano
MedicalResearch.com: What is the background for this study?
Response: In the context of SARS-CoV-2 diagnosis, laboratories play a critical role in confirming the initial clinical suspicion of this disease, as confirmation of SARS-CoV-2 presence is essential to ensure the prompt initiation of containment and treatment protocols. This is of utmost importance to avoid further spread of the pandemic, and to assure the best clinical and therapeutic management of the infected patients in the hospital setting. Unfortunately, currently used rtPCR assays lack of the necessary sensitivity to identify all cases of SARS-CoV-2 infection (20% of false negative results [Li D et all, Korean J Rad 2020; Zhao et all CID 2020]). Complementary laboratory assays are therefore strongly needed. Droplet Digital PCR (ddPCR) is a highly sensitive assay for the direct detection and quantification of DNA and RNA targets. It has been increasingly used in infectious disease settings, especially thanks to its ability to consistently and reliably detect down to few copies of viral genomes. Standing the necessity of a limitation (as much as possible) of false negative results in COVID-19 diagnosis, the use of ddPCR could provide a critical support. In the context of COVID-19 diagnosis, two recent studies highlight the good performances of ddPCR in detecting low viral load samples (Suo T MedRxiv 2020; You F MedRxiv 2020)
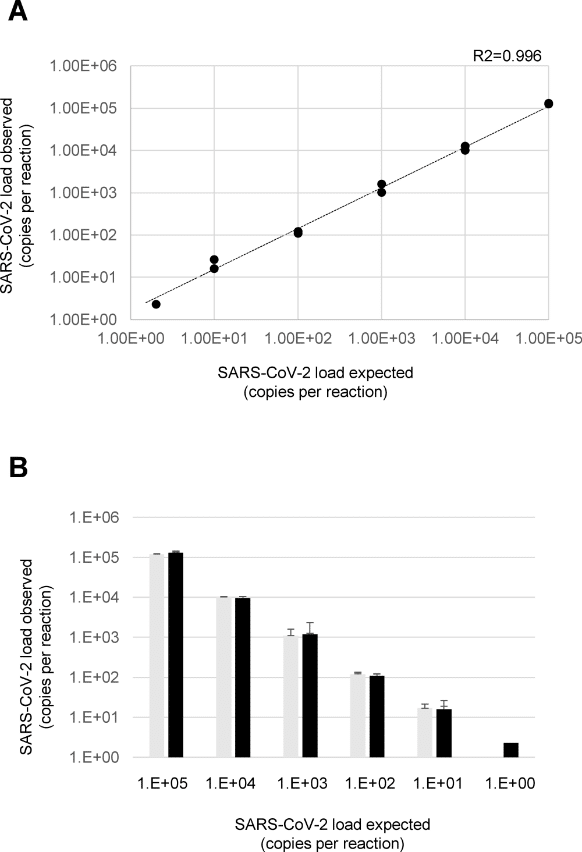
Quantification of SARS-CoV-2 by ddPCR.
A) The graph shows the linear relationship between the expected and the observed concentrations using serial dilutions of the SARS-CoV-2 reference in the two experiments B) The histogram reports the concentrations of the SARS-CoV-2 reference obtained in the first (light grey) and second (dark grey) experiment. In particular, each expected concentration was tested in two independent experiments each led in duplicate (the lowest concentration, 2 copies per reaction, led in quadruplicate). Image: Viecelli et al, 2020 (PLOS ONE, CC BY 4.0
MedicalResearch.com: What are the main findings?
Response: The home-made ddPCR-based assay that we designed targeted the RNA dependent RNA polymerase (RdRp) of SARS-CoV-2 and was able to quantify SARS-CoV-2 copies with a limit of detection of 13 copies per mL of swab. This assay allowed the detection of viral genome in nasopharyngeal samples of 34.5% subjects with a clinical suspect of COVID-19 but with negative RT-PCR standard tests (median viral-load: 128 copies/mL, IQR: 72-345). Serological tests performed at later time points detected SARS-CoV-2 antibodies in all patients tested positive for SARS-CoV-2 in ddPCR (100%), while negative antibody findings were observed in 95.0% of patients tested negative for SARS-CoV-2, thus confirming the good performances of the molecular assay.
MedicalResearch.com: What should readers take away from your report?
Response: ddPCR can be considered as a complement to the current standard rtPCR-based diagnosis, in order to improve as much as possible the rapid identification of SARS-CoV-2 infection (making possible the diagnosis long before the viral load peak is reached and antibodies appear) and thus the definitive containment of the pandemics. Even more relevant, this test could be used for treatment monitoring, or for all supposed COVID-19 patients, who are negative for nasopharyngeal swab nucleic acid tests twice by rtPCR, but at risk of carrying SARS-CoV-2.
MedicalResearch.com: What recommendations do you have for future research as a result of this work?
Response: The use of a highly sensitive home-made assay based on a ddPCR system could improve the efficiency of COVID-19 diagnosis on nasopharyngeal swabs even at a very early stage of viral replication. This assay can be also used to check the presence of viral RNA in anatomical districts where the virus can be present but at lower levels, such as faecal or plasma samples. This provides a significant clinical advantage in terms of prompt identification of subjects with SARS-CoV-2 infection, and for the consequent implementation of all appropriate containment and therapeutic measures. By a clinical research point of view, this test can reveal and accurately quantify the presence of SARS-CoV-2 in new anatomical districts, providing a consistent help in all those clinical scenarios in which is of great importance to identify even the smallest amount of viral particles
MedicalResearch.com: Is there anything else you would like to add?
Response: We thank Bio-Rad Italia for providing technical support, particularly Dr. Laura Sard, Dr. Marilisa Marinelli and Dr. Marco Tronconi and Medical Affairs Bio-Rad team, for their valuable assistance.
Citation:
Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0236311
JOIN OUR EMAIL LIST
[mailpoet_form id="5"]We respect your privacy and will never share your details.
Last Modified: [last-modified]
The information on MedicalResearch.com is provided for educational purposes only, and is in no way intended to diagnose, cure, or treat any medical or other condition. Always seek the advice of your physician or other qualified health and ask your doctor any questions you may have regarding a medical condition. In addition to all other limitations and disclaimers in this agreement, service provider and its third party providers disclaim any liability or loss in connection with the content provided on this website.
Last Updated on September 9, 2020 by Marie Benz MD FAAD
