MedicalResearch.com Interview with:
Professor Anasse Bari PhD
Courant Institute of Mathematical Sciences,
Computer Science Department, New York University, New York, and
Megan Coffee MD PhD
Division of Infectious Diseases and Immunology, Department of Medicine
New York University,
Department of Population and Family Health
Mailman School of Public Health
Columbia University, New York
MedicalResearch.com: What is the background for this study?
Coffee and Bari: This work is led by NYU Grossman School of Medicine and NYU’s Courant Institute of Mathematical Sciences, in partnership with Wenzhou Central Hospital and Cangnan People's Hospital, both in Wenzhou, China. This is a multi-disciplinary team with backgrounds in clinical infectious disease as well as artificial intelligence (AI) and computer science.
There is a critical need to better understand COVID-19. Doctors learn from collective and individual clinical experiences. Here, no clinician has years of experience. All are learning as they go, having to make important decisions about clinical management with stretched resources. The goal here is to augment clinical learning with machine learning.
In particular, the goal is to allow clinicians to identify early who from the many infected will need close medical attention. Most patients will first develop mild symptoms, yet some 5-8 days later will develop critical illness. It is hard to know who these people are who will need to be admitted and may need to be intubated until they become ill. Knowing this earlier would allow more attention and resources to be spent on those patients with worse prognoses. If there were ever treatments in the future that could be used early in the course of illness, it would be important to identify who would most benefit
We present in this study a first step in building an artificial intelligence (AI) framework, with predictive analytics (PA) capabilities applied to real patient data, to provide rapid clinical decision-making support. It is at this point a proof of concept that it could be possible to identify future severity based on initial presentation in COVID-19.
(more…)




 Dr. Helen Marsden PhD
Skin Analytics Limited
London, United Kingdom
MedicalResearch.com: What is the background for this study?
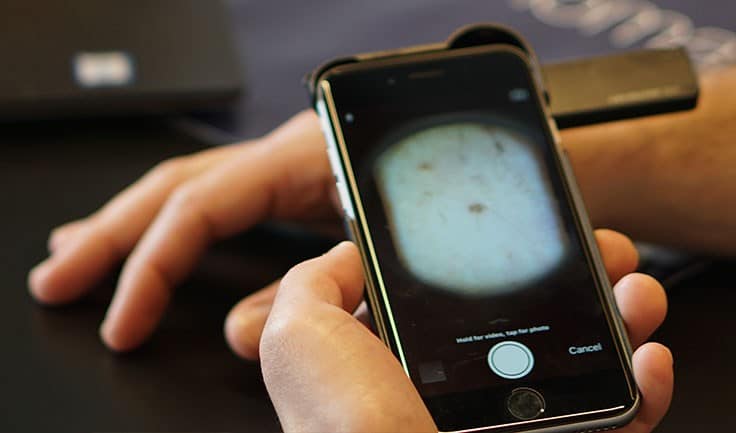
Response: In this technology age, with the explosion of interest and applications using Artificial Intelligence, it is easy to accept the output of a technology-based test - such as a smartphone app designed to identify skin cancer - without thinking too much about it. In reality, technology is only as good as the way it has been developed, tested and validated. In particular, AI algorithms are prone to a lack of “generalisation” - i.e. their performance drops when presented with data it has not seen before. In the medical field, and particularly in areas where AI is being developed to direct a patient’s diagnosis or care, this is particularly problematic. Inappropriate diagnosis or advice to patients can lead to false reassurance, heightened concern and pressure on NHS services, or worse. It is concerning, therefore, that there are a large number of smartphone apps available that provide an assessment of skin lesions, including some that provide an estimate of the probability of malignancy, that have not been assessed for diagnostic accuracy.
Skin Analytics has developed an AI-based algorithm, named: Deep Ensemble for Recognition of Malignancy (DERM), for use as a decision support tool for healthcare providers. DERM determines the likelihood of skin cancer from dermoscopic images of skin lesions. It was developed using deep learning techniques that identify and assess features of these lesions which are associated with melanoma, using over 7,000 archived dermoscopic images. Using these images, it was shown to identify melanoma with similar accuracy to specialist physicians. However, to prove the algorithm could be used in a real life clinical setting, Skin Analytics set out to conduct a clinical validation study.
Dr. Helen Marsden PhD
Skin Analytics Limited
London, United Kingdom
MedicalResearch.com: What is the background for this study?
Response: In this technology age, with the explosion of interest and applications using Artificial Intelligence, it is easy to accept the output of a technology-based test - such as a smartphone app designed to identify skin cancer - without thinking too much about it. In reality, technology is only as good as the way it has been developed, tested and validated. In particular, AI algorithms are prone to a lack of “generalisation” - i.e. their performance drops when presented with data it has not seen before. In the medical field, and particularly in areas where AI is being developed to direct a patient’s diagnosis or care, this is particularly problematic. Inappropriate diagnosis or advice to patients can lead to false reassurance, heightened concern and pressure on NHS services, or worse. It is concerning, therefore, that there are a large number of smartphone apps available that provide an assessment of skin lesions, including some that provide an estimate of the probability of malignancy, that have not been assessed for diagnostic accuracy.
Skin Analytics has developed an AI-based algorithm, named: Deep Ensemble for Recognition of Malignancy (DERM), for use as a decision support tool for healthcare providers. DERM determines the likelihood of skin cancer from dermoscopic images of skin lesions. It was developed using deep learning techniques that identify and assess features of these lesions which are associated with melanoma, using over 7,000 archived dermoscopic images. Using these images, it was shown to identify melanoma with similar accuracy to specialist physicians. However, to prove the algorithm could be used in a real life clinical setting, Skin Analytics set out to conduct a clinical validation study.





